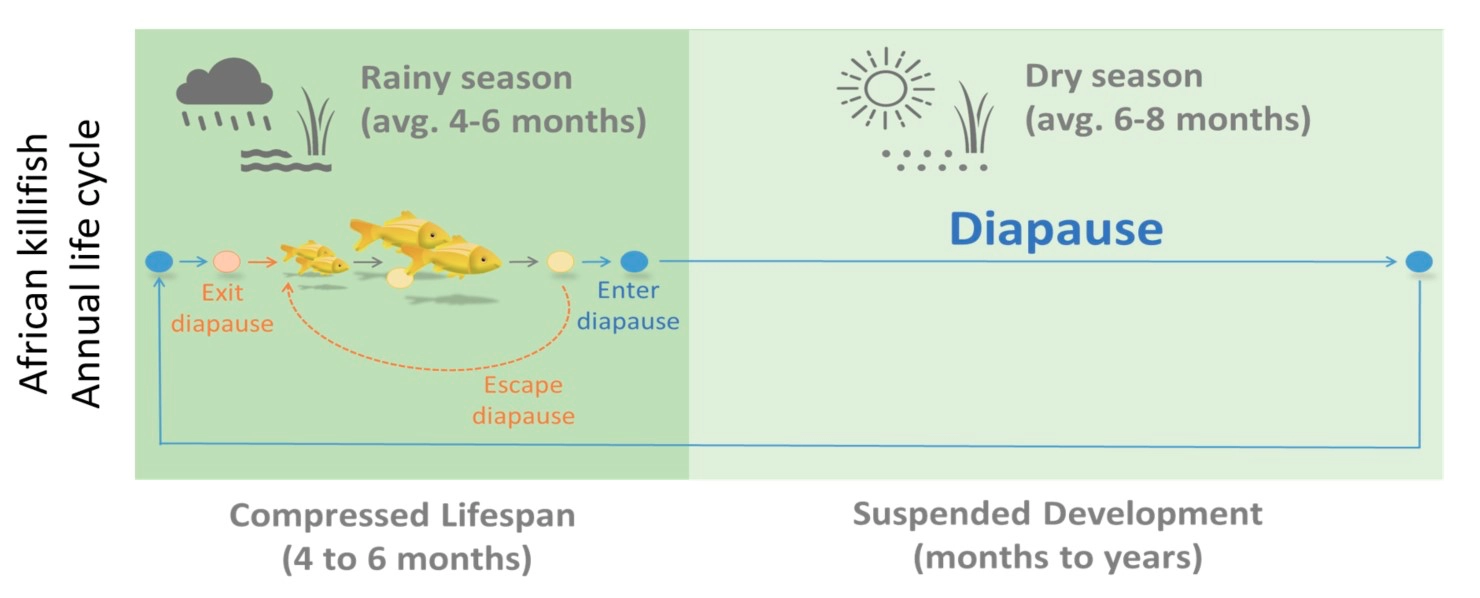
The African turquoise killifish Nothobranchius furzeri (the African killifish hereafter) is a textbook example of species evolving under intense selection pressure to adapt to an extreme environment. The African killifish habituates in ephemeral ponds in Zimbabwe & Mozambique. These seasonal ponds only hold water for a few months during the rainy season and desiccate in the following long dry season. To survive this extreme habitat, the African killifish evolved two remarkable adaptations. First, it grows quickly and starts breeding in 2-3 weeks post-hatching to ensure its propagation within the short rainy season. Then, the majority of newly laid embryos will enter diapause to suspend their development and live through the drought in the form of embryos for months, sometimes even years, in dry sediment until the next rainy season when the rain refills the ponds again. Importantly, these two phases – fast-growing and suspended development – remain unchanged in captivity, even in constant water, indicating that both are under genetic determination.
Consistent with the fast pace of growth in the rainy season, the African killifish also ages fast. Since the African killifish was first found and identified as a new species in 1969, its short-lived nature was broadly known among killifish hobbyists but remained unnoticed to scientists for decades until 2003, when it was first proposed as a research organism for aging. Since then, the increased knowledge and improved husbandry have significantly lowered the early mortality rate and stabilized the median lifespan at 4-6 months, positioning the African killifish as the shortest-lived vertebrate that can breed in captivity. The African killifish recapitulates various stereotypical aging traits of vertebrates, including the decline in reproduction, fertility, cognition, mobility, regeneration, and tissue homeostasis, along with increased incidence of senescence, neural and muscular degeneration, and cancerous lesions, providing a fast and versatile genotype-to-phenotype platform for research.
The African killifish is easy to raise and can live with a broad range of temperature and salinity. In general, any zebrafish systems can be used for the African killifish. The African killifish has the XY sex-determination system and shows sexual dimorphism after maturation, as males are bigger in size and have colorful fin. The African killifish requires no specific conditioning to trigger breeding behaviors. Males and females can be housed in the same tank, breed continuously, and produce tens to hundreds of embryos per week throughout their lives. Due to the African killifish’s fast maturation rate, a stable line with the desired genetic modification can be generated in 2 to 3 months. This is especially advantageous for projects requiring high numbers of modified lines and even more if the time for back-crossing is also considered. Also, the short-lived African killifish is in the sweet spot between canonical invertebrate and vertebrate research organisms. It is an ideal vertebrate platform to test candidates from invertebrates such as C. elegans or Drosophila. At the same time, it can also be employed as a fast-tracked pipeline to functionally verify discoveries from longer-lived vertebrates such as mice, humans, and other long-lived species.
With collective efforts, various types of resources, including genetically diverse strains, genomic datasets, and genome-engineering tools, have been developed to transform the African killifish into a promising research organism from bush to bench. The standard strain that is most frequently used is the GRZ strain, an inbred line that originates from a sample collected in 1970 in the Gona-Re-Zhou National Park in Zimbabwe. Several additional samples have been collected in Mozambique and Zimbabwe. Strains derived from these samples have been used for various studies to identify the genetic architecture underlying differences in these strains, such as color and survival under specific lab environments. Great progress has also been made in developing resource for this fish, including de novo assembled and fully annotated reference genome with numerous transcriptomic and epigenomic datasets, and highly efficient transgenesis and genome editing techniques. Together, the diverse inbred lines and wild strains, the existence of a well-annotated genome, and the ability to manipulate the genome to delete, modify, or overexpress genes, have established this species as a highly promising new research organism in vertebrates.